Comparative Genomics
The evolutionary origin of cnidocytes in the stinging black sponge Haliclona cnidata
Institution: Justus Liebig University Giessen
They are understated yet highly successful – marine sponges are evolutionary masters of defense and typically possess high levels of bioactivity to deter predators and competitors. In the case of the stinging black sponge (Haliclona cnidata), these mechanisms are complemented by “cellular weapons” in the form of functional cnidocytes (see Figure 1), usually known only from cnidarians such as jellyfish or corals.
The origin of these cnidocytes is still largely unknown. The sponge might take up the cells by kleptocnidism (translated as the clawing of cnidocytes). This involves taking up and storing cnidocytes or their precursor stages from other cnidarians such as corals in the environment. Alternatively, it may live in a symbiotic relationship with cnidarians that find shelter in its tissues and, in turn, produce and provide cnidocytes to it. Or it has even incorporated genes from the cnidarians into its own genome and can now build these cells on its own.
In our research project “The evolutionary origin of cnidocytes in the stinging black sponge Haliclona cnidata” we want to understand the origin of cnidocytes using experimental laboratory research and genome analyses in the Department of Animal Ecology and Special Zoology at the Justus Liebig University Giessen (JLU).
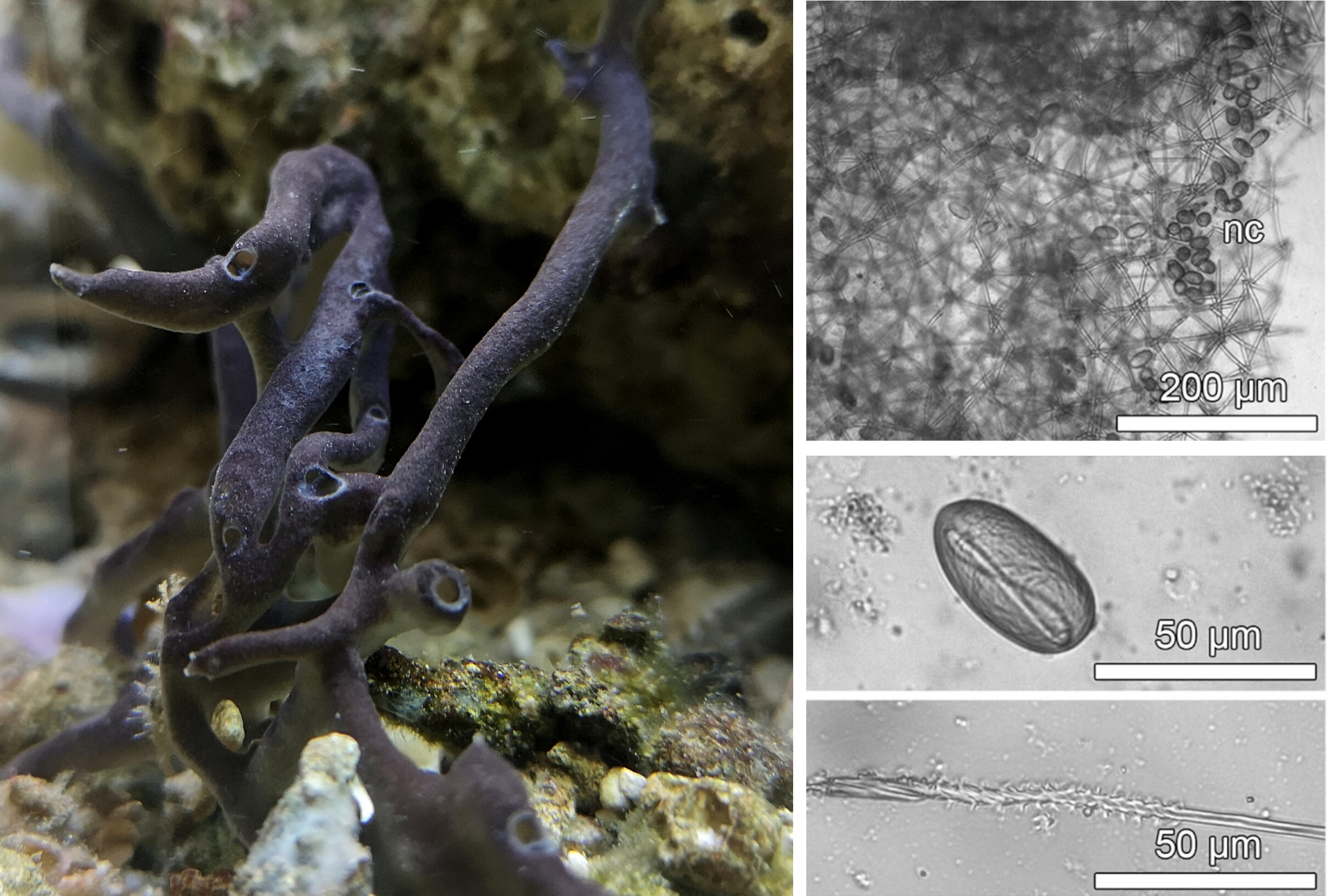
Figure 1: Sponges occur in marine habitats as significant members of benthic environments. The stinging black sponge (Haliclona cnidata) is easy to overlook and has so far only been found in tropical marine aquaria (left). For its defense, like cnidarians, it has cnidocytes (nc) stored in the outer layer of its tissue, which are triggered upon distress and can inject a highly effective toxin into its attackers (right).
Group expertise, Methods
- Structure and function of metaorganisms
- Model organisms
- Symbiosis
- Microbial community analyses
- Morphometrics
- Experimental marine biology
- Simulations of global change
- Ecophysiology
Genome data
The project will involve sequencing the genome of the sponge. The bioinformatic analysis will identify a large number of microbial symbionts in addition to the sponge. As sessile filter feeders, sponges are constantly exposed to environmental microorganisms that can be both beneficial and harmful to the sponge. Haliclona cnidata also belongs to the “high microbial abundance” (HMA) sponges and harbors a complex, sponge-specific and evolutionarily adapted microbial community (Figure 2) which together with the sponge forms a functional unit – the sponge holobiont. In the next steps, we plan to analyze the sponge isolated from its symbionts to provide a better data basis for bioinformatic analyses. In experimental laboratory studies the formation of the cnidocytes under controlled conditions will be investigated in more detail.
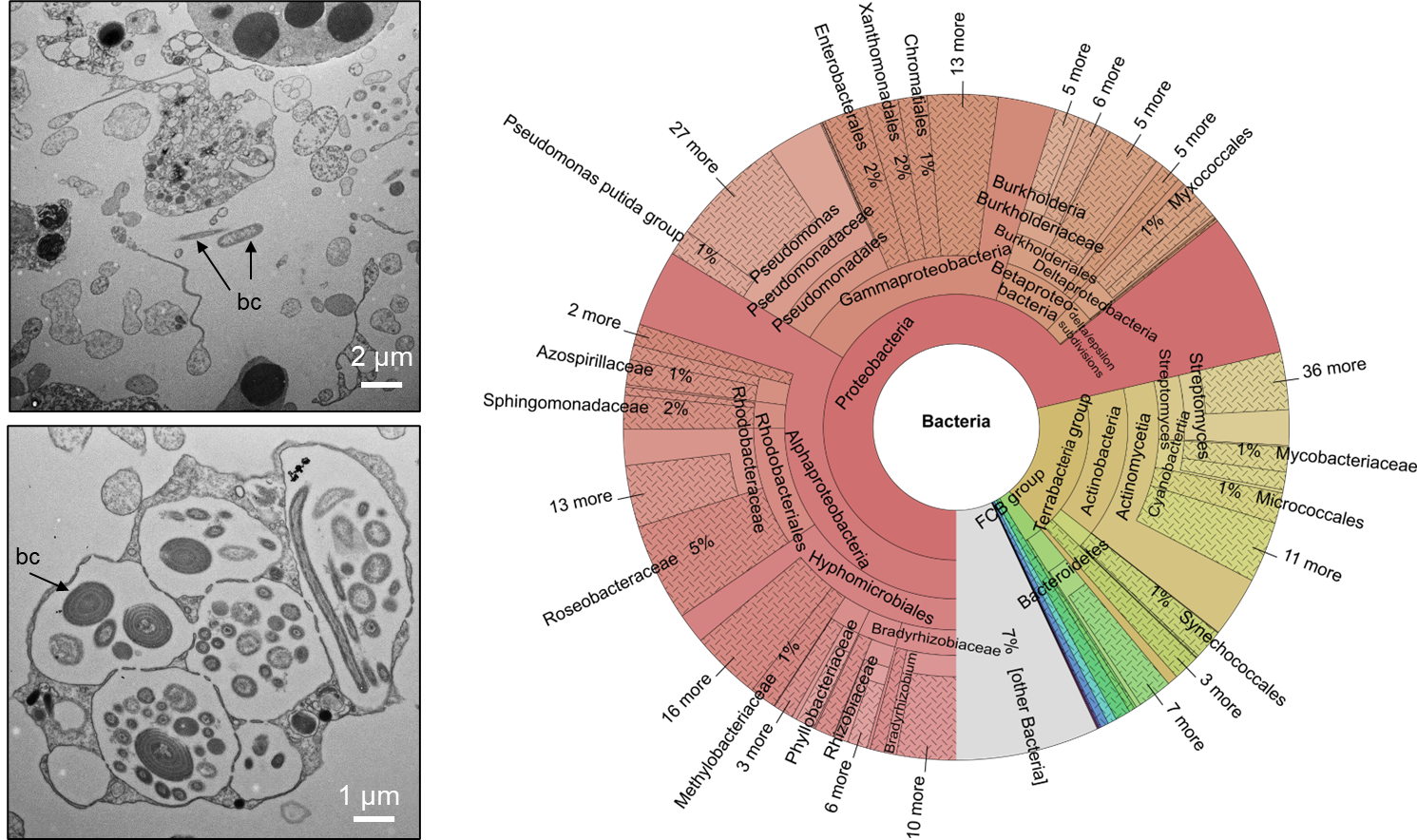
Figure 2: Bacteria (bc) occasionally live in the mesohyl (top left) and more often in specialized compartments called bacteriocytes in the stinging black sponge, as seen here in a transmission electron microscopy (TEM) image (bottom left). The complex bacterial community was analyzed in a first study based on a genome sequencing of the holobiont (right). Source: Schellenberg et al. 2020.
The sponge was first analyzed and described as part of the doctoral thesis of Johannes Schellenberg at the Institute of Animal Ecology and Special Zoology at the Justus Liebig University in Giessen. First genome analyses were performed as part of the master thesis of Yvonne Pfeifer in cooperation with Jörn Kalinowski and Daniel Wibberg from CeBiTec in Bielefeld. Sponge tissue of the sponge holobiont was used to assemble the genome using bioinformatic tools (BUSCO, Kraken2) and to characterize the symbiont community of the sponge.
The analysis of the sponge is a collaborative project, taking place in the “Ocean2100” research facility at the University of Giessen. The “Ocean2100” facility is funded by the CEMarin (Center of Excellence in Marine Sciences) and can simulate global change scenarios in the ocean to test their effects on stony corals and other associated reef organisms.
Selected publications
Schellenberg, J., Busse, H. J., Hardt, M., Schubert, P., Wilke, T., Käampfer, P., & Glaeser, S. P. (2018). Proposal of Litorimonas haliclonae sp. Nov., isolated from a marine sponge of the genus Haliclona. International Journal of Systematic and Evolutionary Microbiology, 68(3), 835–843. https://doi.org/10.1099/ijsem.0.002592
Schellenberg, J., Reichert, J., Hardt, M., Klingelhöfer, I., Morlock, G., Schubert, P., Bižić, M., Grossart, H. P., Kämpfer, P., Wilke, T., & Glaeser, S. P. (2020). The Bacterial Microbiome of the Long-Term Aquarium Cultured High-Microbial Abundance Sponge Haliclona cnidata – Sustained Bioactivity Despite Community Shifts Under Detrimental Conditions. Frontiers in Marine Science, 7(May). https://doi.org/10.3389/fmars.2020.00266
Schellenberg, J., Reichert, J., Hardt, M., Schmidtberg, H., Kämpfer, P., Glaeser, S. P., Schubert, P., & Wilke, T. (2019). The precursor hypothesis of sponge kleptocnidism: Development of nematocysts in Haliclona cnidata sp. nov. (porifera, demospongiae, haplosclerida). Frontiers in Marine Science, 5(JAN), 1–14. https://doi.org/10.3389/fmars.2018.00509