TBG Laboratory
Dr. Carola Greve, Leonie Schardt, Damian Baranski, and Alexander Ben Hamadou
Together with the bioinfomatics service unit the TBG laboratory has established a data management system that organizes and stores the raw and meta data of >3000 Sequencing runs (as of 08/2021) to make them available for all PIs at TBG and prepare submission of the data.



We are well equipped to master the challenges to isolate hmwDNA from all corners of life, ranging from mammals, molluscs and insects over to plants, lichens and fungi. As PacBio beta tester we sequence entire de novo genomes from tiny organisms (1-2 mm) with as little as 5 ng total input gDNA. Our equipment allows us to make all necessary quality checks including genome size estimation, pulse-field electrophoresis, as well as fragmentizing to produce ready–to-use libraries that we sequence at various collaborators (BGI, Novogene, Radboud University Medical Center, SciLife). Some sequencing is done in-house on Nanopore (about 60 genomes) or iSeq 100 for quality testing and small projects.
Equipment
- Pulsfeld Elektrophorese (Analytik Jena)
- 2200 TapeStation (Agilent Technologies)
- Qubit 2.0 (Life Technologies)
- BluePippin (Sage Science)
- MinION (Oxford Nanopore)
- Illumina Next Generation Sequencing (NGS) platform iSeq 100
- Bioruptor ® Pico (Diagenode)
- CytoFLEX Flow Cytometer (Beckman Coulter)
- Femtopulse (Agilent)
- Bioanalyzer (Agilent)
- Plate reader (M2, Molecular devices)
- qPCR-Maschine (CFX Opus 96, Bio-Rad)
- 2x Thermocycler (Mastercycler X50t, Eppendorf)
- Pipetting robot
M220 Focused-ultrasonicator
GENOMES SEQUENCED
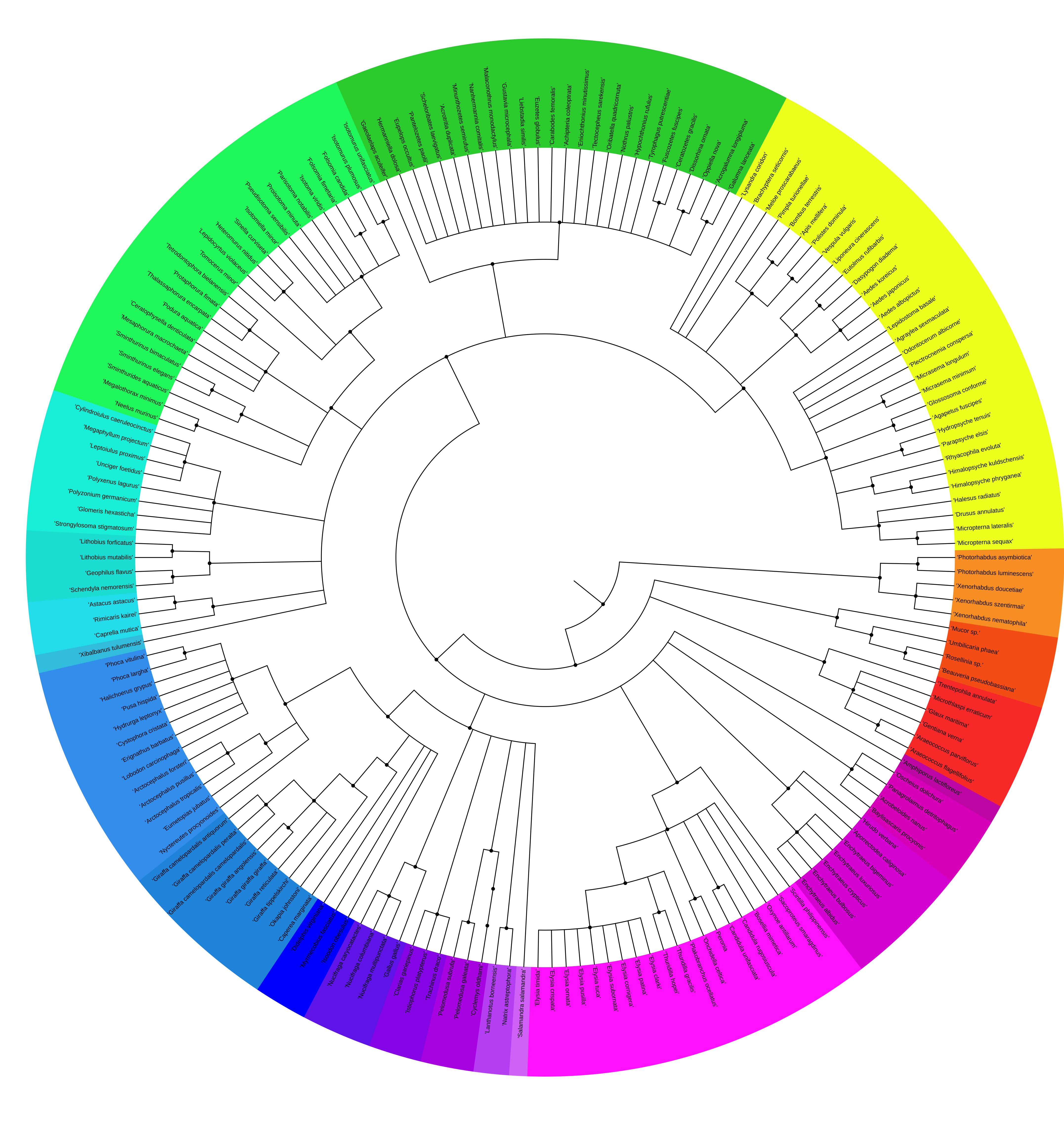
Overview of species sequenced by TBG: > 351 species from 11 phyla
TBG Lab Center Team

Dr. Carola Greve (Lab and Project management)
carola.greve@senckenberg.de
Leonie Schardt (Labmanager FUG “Functional Environmental Genomics”)
leonie.schardt@senckenberg.de
Damian Baranski (Labmanager Robotics FUG “Functional Environmental Genomics” und GBM “Genomic Biomonitoring”)
damian.baranski@senckenberg.de
Alexander Ben Hamadou (Technical Assistant)
alexander-ben.hamadou@senckenberg.de
Charlotte Gerheim (Technical Assistant)
charlotte.gerheim@senckenberg.de