Comparative Genomics
Unraveling the genetic basis of functional kleptoplasty in "solar-powered" sea slugs
Institution: Senckenberg Society for Nature Research
“Solar-powered” sea slugs are fascinating! They belong to the group Sacoglossa (Heterobranchia) and have the ability to incorporate chloroplasts from their food algae. These “stolen” plastids (kleptoplasts) are then stored in the slug’s digestive gland cells and continue photosynthesis inside the slugs. This phenomenon is called functional kleptoplasty and has only been observed in animals of the group Sacoglossa and in some marine flatworms. To date, the genetic basis that enables this lifestyle is poorly understood.
Our project involves the functional evaluation of DNA methylation during the life cycle of Elysia timida, which can store chloroplasts of the green alga Acetabularia acetabulum for up to 6 months. Epigenetic mechanisms regulate the interpretation of genetic information and adapt gene expression patterns to changing developmental or environmental conditions. Several epigenetic mechanisms have been identified to date, with DNA cytosine methylation being the best studied and possibly the most important epigenetic mark. In this project, we will compare gene expression and DNA methylation patterns in newly hatched individuals that have not yet incorporated chloroplasts with those of adult individuals that have already incorporated chloroplasts with their food algae. By analyzing experimental time series, this project will represent the first high-resolution characterization of DNA methylation patterns and gene expression in a “solar-powered” sea slug species to understand the fundamental role of this epigenetic mark in the molluscan phylum (including an investigation of the relationship between gene expression and DNA methylation) and identify potential genes important for successful chloroplast incorporation.
In addition, sacoglossan sea slugs are known to incorporate secondary metabolites from their food algae to become unpalatable to predators (kleptochemistry). In recent years, a number of structurally diverse γ-pyrone containing polypropionates natural products have been isolated from this group. Many of these metabolites possess complex structural frameworks and have been shown to exhibit interesting biological activities. Here, we are working closely with the LOEWE TBG program area Natural Product Genomics (Prof. Dr. Eric Helfrich).
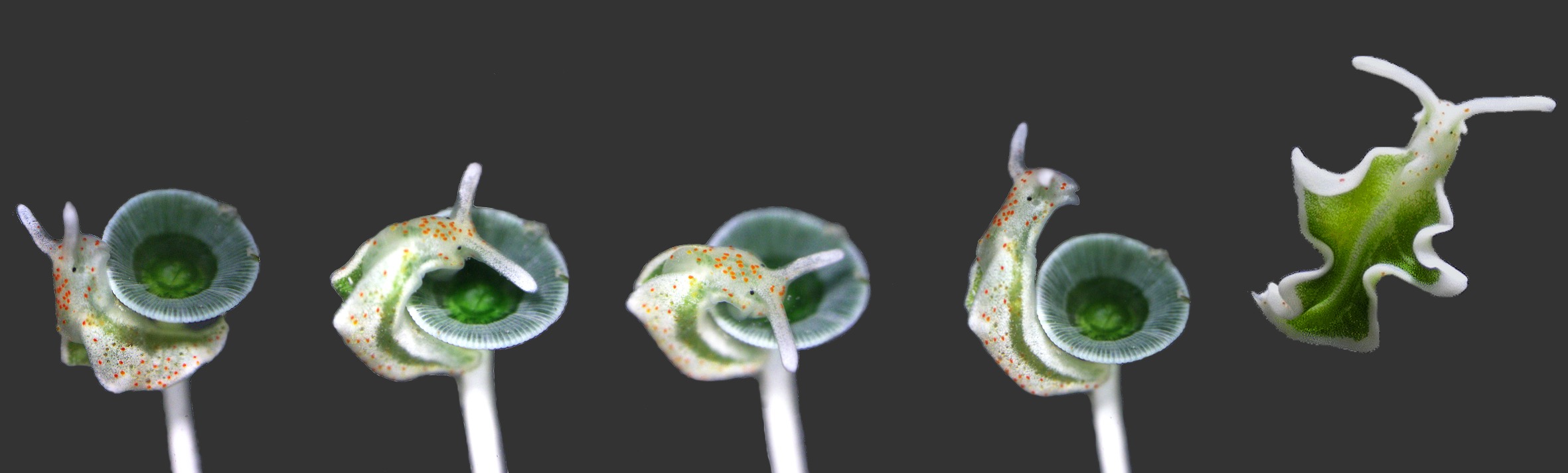
Elysia timida on Acetabularia acetabulum. Fotos: Heike Wägele & Victoria Morris. Collage: Carola Greve
Group members
- Dr. Carola Greve (PI)
- Prof. Dr. Julia Sigwart
- Dr. Tilman Schell (PI)
- Lisa Männer
- Charlotte Gerheim
- Alexander Ben Hamadou
Contributors
- Prof. Dr. Eric Helfrich (LOEWE Centre for Translational Biodiversity Genomics, Natural Product Genomics, Frankfurt, Germany)
- Thao Phan Ngoc (LOEWE Centre for Translational Biodiversity Genomics, Natural Product Genomics, Frankfurt, Germany)
- Dr. Panagiotis Provataris (Heidelberg, Germany)
- Carles Galià-Camps (Universitat de Barcelona, Barcelona, Spain)
Group expertise, Methods
- DNA & RNA extractions from a wide variety of organisms
- Preparation of NGS libraries (PacBio, Nanopore, Hi-C, Samplix)
- Genome sequencing with PacBio and Illumina
- RNAseq & Iso-Seq (PacBio)
- De novo assembly and annotation of whole genomes
- De novo transcriptome assembly
- Chromosome-scale scaffolding (Hi-C)
- Comparative genomics
- DNA methyation
Genomes sequenced
- Elysia timida
- Elysia cornigera
- Elysia subornata
- Thuridilla hopei
- Ercolania ermarginata
Selected publications
- Männer, L., Schell, T., Provataris, P., Haase, M., & Greve, C. (2021). Inference of DNA methylation patterns in molluscs. Philosophical Transactions of the Royal Society B, 376 (1825), 20200166