Comparative Genomics
Evolution and function of species interaction networks on the example of pioneering species
Institution: Department of Applied Bioinformatics, Goethe University, Frankfurt
TBG GROUP MEMBERS
- Prof. Dr. Ingo Ebersberger
- Dr. Francisca Segers
- Andreas Blaumeiser
- Julian Dosch
TBG PROJECT SUMMARY
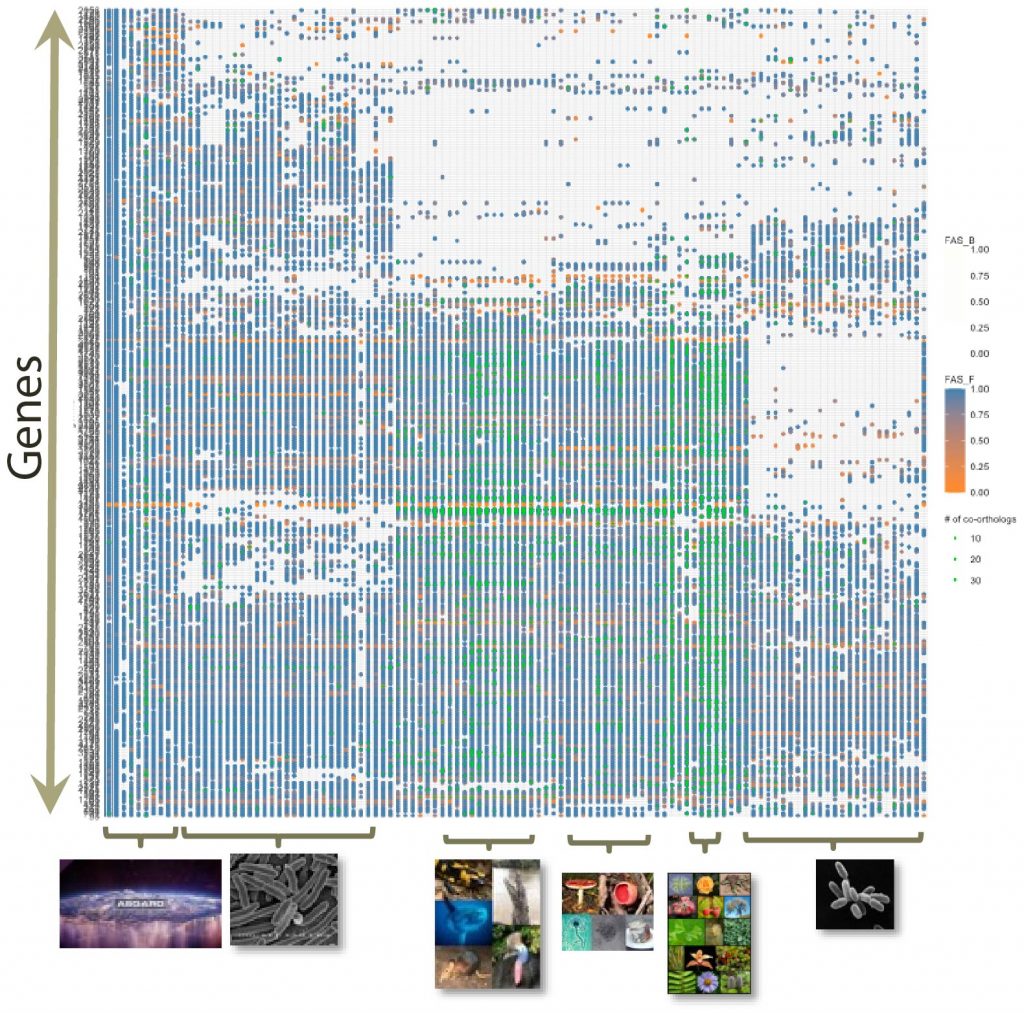
Symbiosis is an evolutionarily successful strategy to optimally exploit resources in a habitat. However, we are only at the beginning of comprehending the molecular basis of organismic interactions. In this project, we combine methods development with applied analysis. Specifically, we will create a bioinformatics toolkit to trace protein interaction networks across many taxa. The novel software will inform about lineage specific changes in protein function and will assess the effect of limited search sensitivity. The applied part of our project concentrates on pioneering communities, i.e. meta-organisms that are the first to conquer new, and often extreme, habitats, such as saline and nutrient-poor soils. Using newly determined genome sequences of symbionts, we will identify encoded protein interaction networks, and their integration across symbiotic partners. Comparisons with results from closely related non-pioneering taxa will shed light on how species adapt to extreme environments
HIGHLIGHTS
GROUP EXPERTISE / METHODS
- Phylogenomics
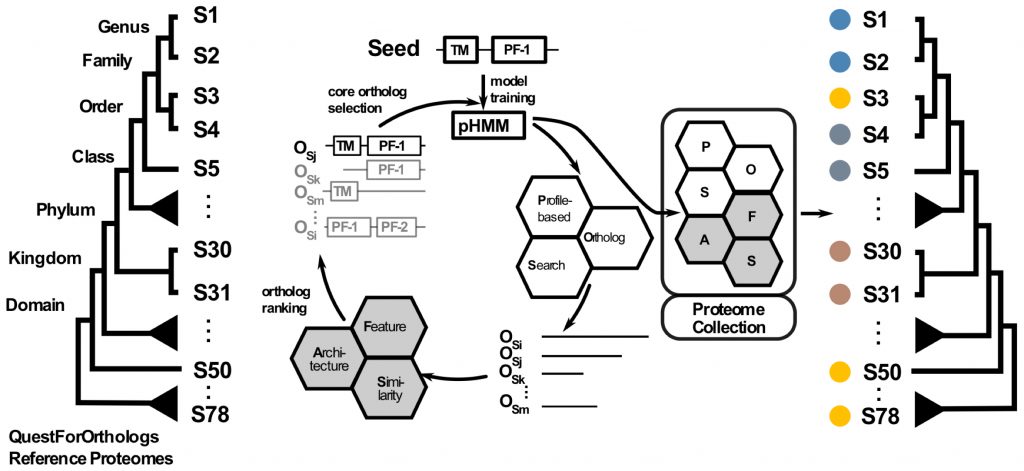
- Orthology inference
- Genome assembly
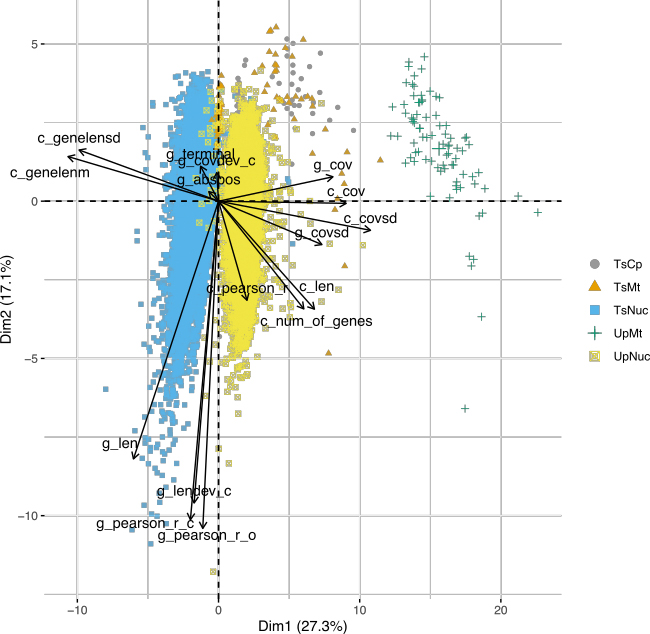
- Comparative genomics
- Microbiome analysis
- General bioinformatics
- Evolutionary biology
- Behavioural ecology
GENOMES ANALYZED
1 — Lichen hologenome Illumia 120x, PacBio 30x
1 — Plant de novo PacBio 50x, Illumina 50x
De novo genomes: 3
Species:
Umbilicaria pustulata
Glaux maritima
Ardisia venosa
Lysimachia punctata
Cyclamen purpurascens
PUBLICATIONS
Comparative transcriptomics reveal insights into the drought response of the three Panicum species P. bisulcatum, P. laetum and P. turgidum
Kotrade, P, et al.
Plant Gene Volume 23, September 2020, https://doi.org/10.1016/j.plgene.2020.100232
Adjustment of photosynthetic activity to drought and fluctuating light in wheat
Grieco, M. et al. Plant Cell Environ. 2020;1-17; https://doi.org/10.1111/pce.13756
What is in Umbilicaria pustulata? A metagenomic approach to reconstruct the holo-genome of a lichen
Greshake Tzovaras, B.; Segers, F.; Bicker, A.; Dal Grande, F.; Otte, J.; Yahya Anvar, S.; Hankeln, T.; Schmitt, I.; Ebersberger, I.;
Genome Biology and Evolution 2020, eva0049
Advances and Applications in the Quest for Orthologs.
Glover N, et al.
Mol Biol Evol. 2019 Oct 1;36(10):2157-2164. doi: 10.1093/molbev/msz150.#
The evolutionary traceability of a protein
Jain, A. et al.
Genome Biol Evol. 2019 Feb 1;11(2):531-545. doi: 10.1093/gbe/evz008.