Comparative Genomics
Strategies to optimize the analysis of gene flow in speciation at the example of giraffes, bears, whales, and seals
Institution: Senckenberg Gesellschaft für Naturforschung
SUMMARY
In this project, we are sequencing mammalian genomes to study how species differ at the sequence level. In particular methods implemented in D-stat, Phylonet, D-foil, CoalHMM show how past and current gene flow affects speciation. Initially, we will focus on current projects in giraffe, bears and seals. For these taxonomic groups geneflow has been documented or is suspected from small scale studies. We are sequence entire genomes, often in a population genomic approach, to study the extend of geneflow arcoss the genome. Alternative presentations such as networks and circus plots will depict of evolutionary processes. The expected inflation of the number of genomes for numerous species will make it necessary to develop efficient procedures to involve hundreds of species/individuals. Due to changes of the PI, this project is behind schedule, but 50 giraffe genomes are sequenced and I gathered an international consortium to study geneflow in bears across their distribution range
Highlights
Migration rates among four giraffe clades:
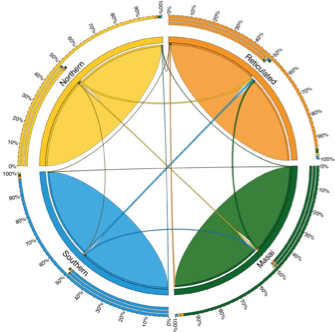

Circular migration plot of recent migration rates among four giraffe clades estimated by BayesAss and indicated by ribbons connecting one species to another. The color coding represents the four species.
Geneflow signals for baleen whales:

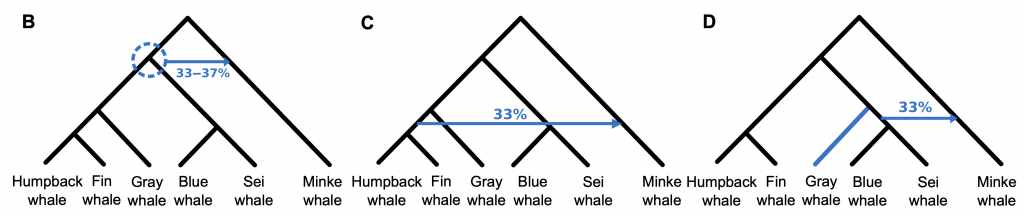
Geneflow signals for baleen whales inferred by PhyloNet.( Rooted networks with reticulations inferred from PhyloNet based on 34,192 20-kbp genome fragments. Reticulations are shown as blue arrows with inheritance probability denoted above or below.
TBG group members
- Prof. Dr. Axel Janke (PI)
- Dr. Menno de Jong
- Dr. Bruno Ferrette
- MSc Sven Winter
- MSc Raphael Coimbra
- MSc Magnus Wolf
- MSc Marcel Nebenfuhr
- MSc David Prochotta
Genomes Sequenced
2 – Giraffes de novo PacBio+OMNI-C+Illumina
1 – Okapi de novo PacBio+OMNI-C+Illumina
450 – Giraffes Illumina 15x
6 – Rorquals de novo PacBio and 10XGenomics
50 – Fin whales Illumina 20x
40 – Blue whales Illumina 20x
250 – Brown bears Illumina 20x
150 – Red deer Illumina 20x
50 – Fanfish Illumina 20x
5 – various fish species de novo PacBio+OMNI-C+Illumina
6 – Baikal seals Illumina 20x- Giraffes de novo PacBio+OMNI-C+Illumina
1 – Okapi de novo PacBio+OMNI-C+Illumina
450 – Giraffes Illumina 15x
6 – Rorquals de novo PacBio and 10XGenomics
50 – Fin whales Illumina 20x
40 – Blue whales Illumina 20x
250 – Brown bears Illumina 20x
150 – Red deer Illumina 20x
50 – Fanfish Illumina 20x
5 – various fish species de novo PacBio+OMNI-C+Illumina
6 – Baikal seals Illumina 20x
Group expertise / Methods
Genome assembly tool
Evolutionary network analyses
Gene flow D-stat and it derivatives
PUBLICATIONS
Coimbra et al “Whole-genome analysis of giraffe supports four distinct species”. Current Biology, Vol. 31, Issue 13,12 July 2021
Stefan Prost, Sven Winter, Jordi De Raad, Raphael T F Coimbra, Magnus Wolf, Maria A Nilsson, Malte Petersen, Deepak K Gupta, Tilman Schell, Fritjof Lammers, Axel Janke, Education in the genomics era: Generating high-quality genome assemblies in university courses, GigaScience, Volume 9, Issue 6, June 2020,
Sven Winter, Stefan Prost, Jordi De Raad, Raphael T. F. Coimbra, Magnus Wolf (…) Maria A. Nilsson, Axel Janke, Chromosome-level genome assembly of a benthic associated Syngnathiformes species: the common dragonet, Callionymus lyra,Gigabyte,1,2020
Árnason, Úlfur, et al. “Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow.” Science advances 4.4 (2018)
TBG GENOMIC STATS:
| De novo genomes | 3 |
| Re- sequenced genomes | 6 |
| Total genomes: | 9 |
| Total data: | 0,751 Tera bases |
SPECIES INVOLVED:
| SPECIES: | Date (due) | NCBI | Coverage Quality (planned) | |
| Giraffes: | 01.01.18 | Yes | 15x | re-sequence |
| Northern giraffe (Giraffa camelopardalis) Nubian | 01.01.19 | No | (15x) | re-sequence |
| Northern giraffe (G. camelopardalis) Rothschild’s | 01.01.19 | No | (15x) | re-sequence |
| Northern giraffe (G. camelopardalis) West African | 01.01.19 | No | (15x) | re-sequence |
| Reticulated giraffe (G. reticulata) | 01.01.19 | No | (15x) | re-sequence |
| Masai giraffe (Gi. tippelskirchi) Masai | 01.01.19 | No | (15x) | re-sequence |
| Masai giraffe (G. tippelskirchi) Luanga Valle NP | 01.01.19 | No | (15x) | re-sequence |
| Southern giraffe (G. giraffa) Angolan | 01.01.19 | No | (100x) | de-novo (gold) |
| Southern giraffe (G.a giraffa) South African | 01.01.19 | No | (15x) | re-sequence |
| Bears: | ||||
| Syrian brown bear (Ursus arctos syriacus) male | 01.07.18 | No | (20x) | re-sequence |
| Syrian brown bear (Ursus arctos syriacus) female | 01.07.18 | No | (20x) | re-sequence |
| Kamchatka brown bear (Ursus arctos beringianus) | 01.07.18 | No | (20x) | re-sequence |
| Wales: | ||||
| NA right whale (Eubalena glacialis) | 06.04.18 | Yes | (10x) | re-sequence |
| Sei whale (Balaenoptera borealis) | 06.04.18 | Yes | (10x) | re-sequence |
| Sei whale (Balaenoptera borealis) | 06.04.18 | Yes | (10x) | re-sequence |
| Blue whale (Balaenoptera musculus) | 06.04.18 | Yes | (30x) | re-sequence |
| Humpback whale (Megaptera novaeangliae) | 06.04.18 | Yes | (10x) | re-sequence |
| Fin whale (Balaenoptera physalus) | 06.04.18 | Yes | (100x) | de-novo (gold) |
| Minke whale (Balaenoptera acutorostrata) | 06.04.18 | Yes | (10x) | re-sequence |
| Gray whale (Eschrichtius robustus) | 06.04.18 | Yes | (10x) | re-sequence |
| Gray whale (Eschrichtius robustus) | 06.04.18 | Yes | (10x) | re-sequence |
| Hippopotamus (Hippopotamus amphibius) | 06.04.18 | Yes | (60x) | de-novo gold |