Comparative Genomics
TEology – automatic detection of transposable elements in assembled eukaryotic genomes
Institution: Senckenberg Society for Nature Research
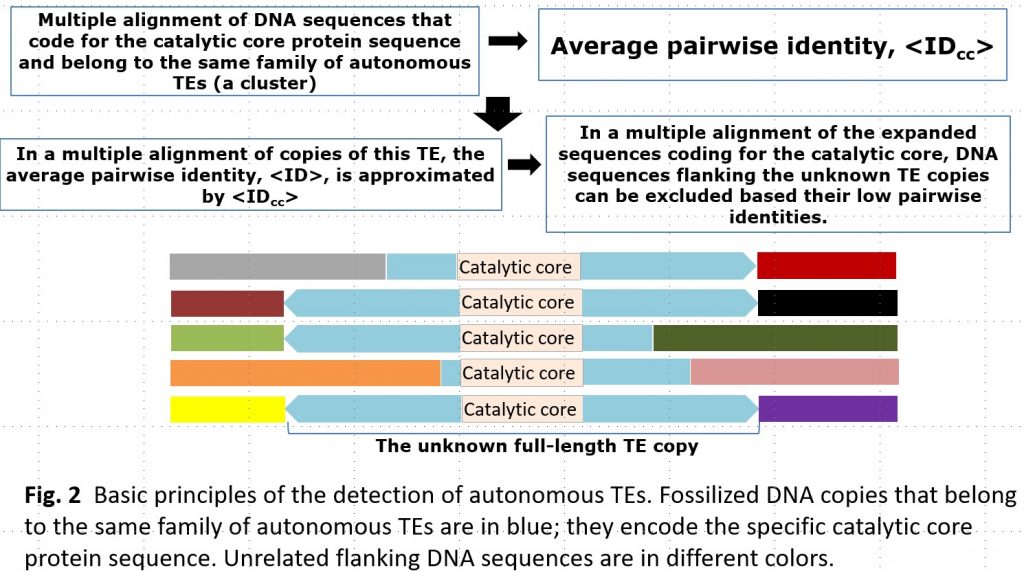
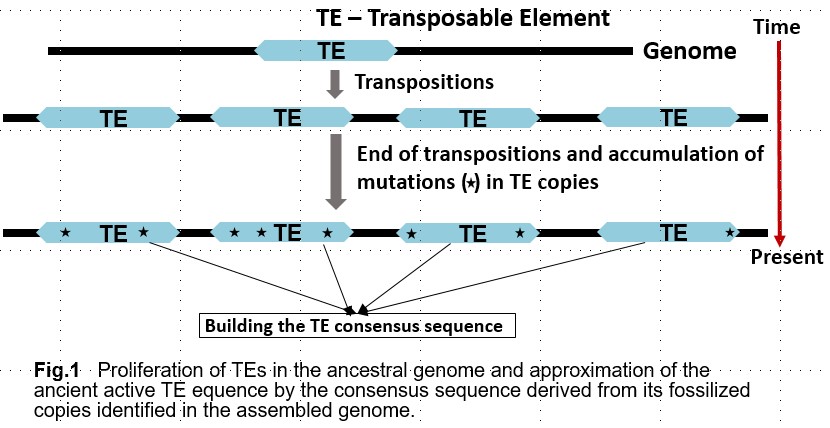
Transposable elements (TEs) are an essential component of the eukaryotic genome and are a major factor of its evolution. In addition to understanding the biological significance of TEs, their accurate detection in a genome is also important for gene prediction and comparative genomics. The exponential growth of the number of assembled genomes is a challenge for manual or semi-automatic detection of TEs. Typical annotation of TEs in the eukaryotic genome is laborious and requires special experts in TEs.
Currently available software, including RepeatModeler, is not accurate enough to identify all TEs, especially in poorly studied genomes. As a result, a large proportion of identified transposable elements (TEs) remain unclassified and undetected. This makes it difficult to investigate the cause of the proliferation of these unclassified TEs. In many cases, the proliferation of TEs is associated with speciation processes and can be attributed to dramatic changes in transposition rates in functional regions, for example, gene transcription promoters, caused by changes in the environment. To better understand the occurrence and nature of these events, we need to know which proteins were involved in the transposition of the unclassified elements.
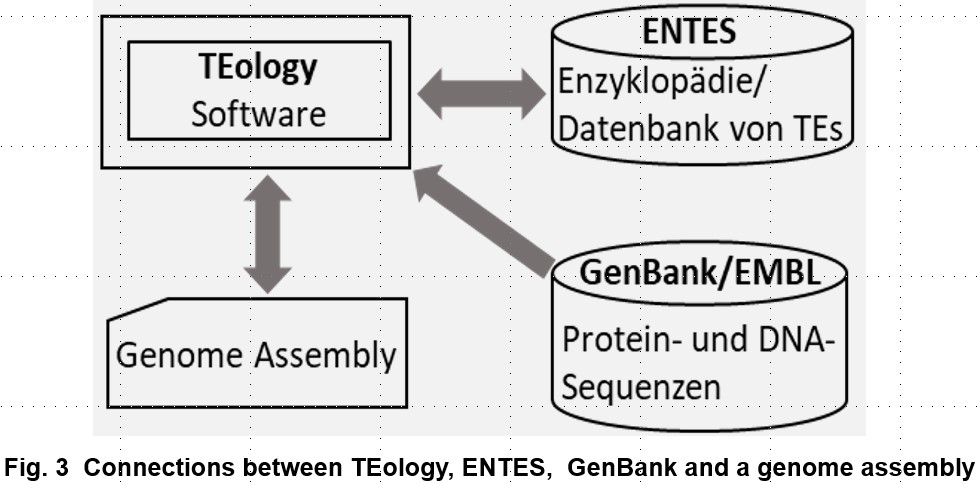
The main goal of the TEology project is to establish a computational pipeline for de novo detection and annotation of TEs in assembled eukaryotic genomes for widely use by TBG/SGN and international community.
To achieve the main goal, we pursue three specific goals in the developmental phase: (a), accurate and fast automatic identification and classification of autonomous and non-autonomous TEs, and discoveries of novel types of autonomous TEs; (b), TEology will be independent of existing TE databases; (c), develop ENTES, an encyclopedia/database of TEs that grows automatically.
PROJECT tEAM
Project PI
Vladimir Kapitonov