Comparative Genomics
Evolution of silk biosynthesis and associated behavior in caddisflies
Foto: Wolfgang Graf
Institution: Senckenberg Gesellschaft für Naturforschung
SUMMARY
Caddisfly larvae use silk in many different ways, e.g. as underwater life lines, to build filtering nets or living retreats, and even to “glue together” mineral or organic particles to build portable cases. This diverse net- and case-making behavior allows them to exploit a range of ecological niches. The properties of caddisfly silk (polymerizes in aquatic environments, high tensile strength and extensibility) make it an interesting material for potential application, e.g. in surgery.
In the first phase, (1) we sequenced and assembled more than 20 genomes from different caddisfly species, and discovered that case-making species have significantly larger genomes compared to net-building species, which may be related to functional diversification and more complex use of silk, (2) the drivers of genome size variation were identified, (3) structural differences in the important L- and H-fibroin silk genes were characterized and a comparison of the silk genes of caddisflies and their terrestrial sister group the Lepidoptera was carried out.
In the next phase, we investigate the evolution and characterization of L and H fibroin genes as central functional elements of the underwater silk of caddisflies and other aquatic insects. The application of the results, the recombinant expression of L and H fibroin genes, will be investigated in collaboration with Prof. Dr. Philipp Seib and collaborators from the Fraunhofer Institute for Molecular Biology and Applied Ecology IME.



Gallery Item Name
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Quidem, provident.

Gallery Item Name
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Quidem, provident.
Gallery Item Name
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Quidem, provident.

Gallery Item Name
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Quidem, provident.

Gallery Item Name
Lorem ipsum dolor sit amet, consectetur adipisicing elit. Quidem, provident.
Comparison of the major silk protein, h-fibroin across caddisfly (Trichoptera) suborders
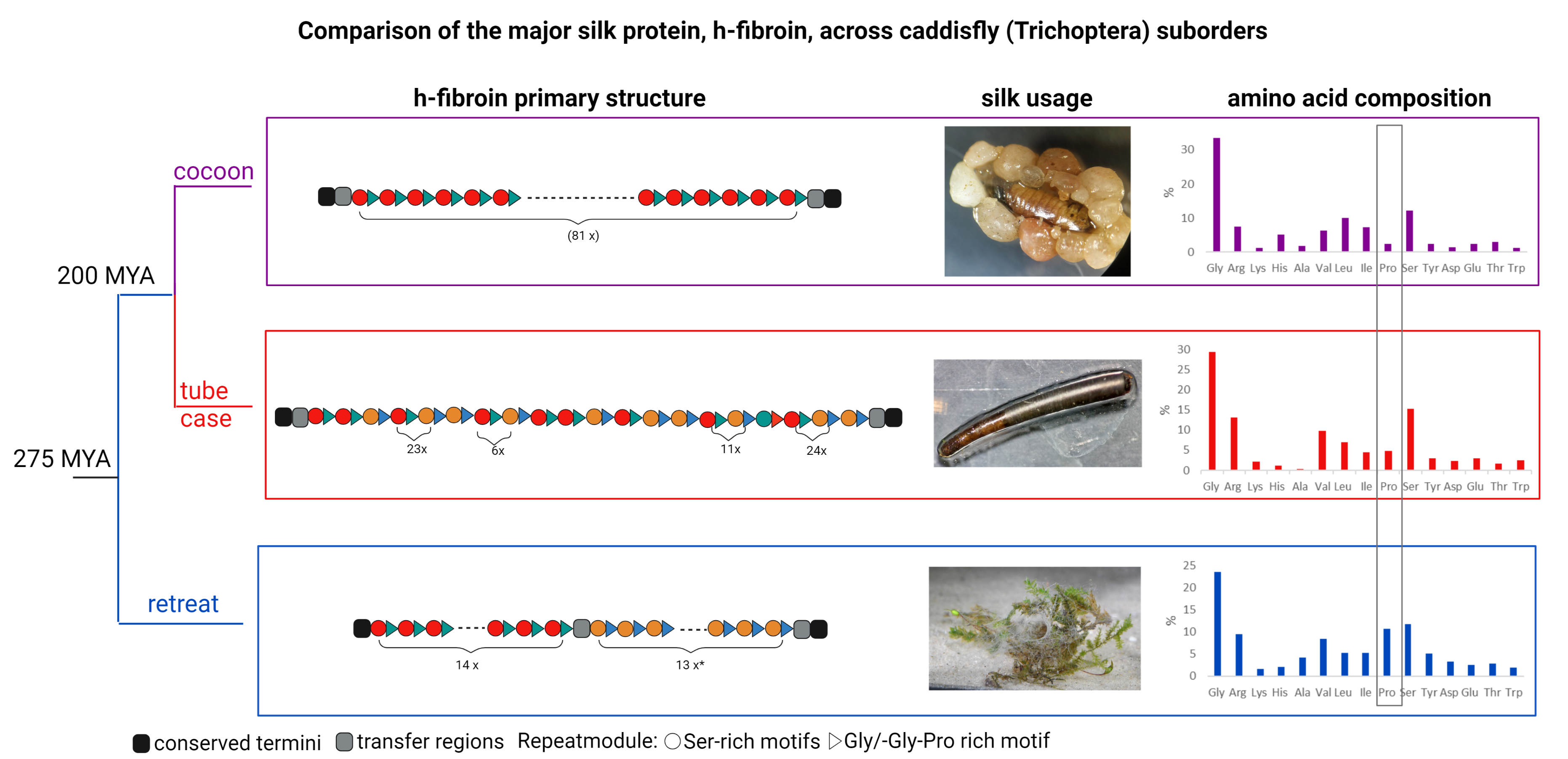
The h-fibroins have conserved termini and basic motif structure with high variation in repeating modules as well as variation in the percentage of amino acids, mainly proline. This finding might be linked to differences in mechanical properties related to the different silk usage and sets a starting point for future studies to screen and correlate amino acid motifs and other sequence features with quantifiable silk properties.
TBG group members
- Prof. Dr. Steffen Pauls (PI)
- Jacqueline Heckenhauer, Ph.D.
- Xiling Deng
- Dr. Julio Schneider
- external: Prof. Paul Frandsen (Brigham Young University)
Group expertise / Methods
- whole genome de novo assembly and annotation
- population genetics
- phylogenetics
- anchored hybrid enrichment



Fotos: Sven Tränkner
Genomes Sequenced
25 de novo genomes of different caddisfly species (Trichoptera).
PUBLICATIONS
Jacqueline Heckenhauer, Russell J. Stewart, Blanca Ríos-Touma, Ashlyn Powell, T. Dorji, Paul B. Frandsen, Steffen U. Pauls (2023). Characterization of the primary structure of the major silk gene, h-fibroin, across caddisfly (Trichoptera) suborders. iScience 26 (8). https://www.sciencedirect.com/science/article/pii/S2589004223013305
Paul B Frandsen, Scott Hotaling, Ashlyn Powell, Jacqueline Heckenhauer, Akito Y Kawahara, Richard H Baker, Cheryl Y Hayashi, Blanca Ríos-Touma, Ralph Holzenthal, Steffen U Pauls, Russell J Stewart (2023). Allelic resolution of insect and spider silk genes reveals hidden genetic diversity, PNAS 120 (18): e2221528120.
Scott Hotaling, Edward R Wilcox, Jacqueline Heckenhauer, Russell J Stewart, Paul B Frandsen (2023). Highly accurate long reads are crucial for realizing the potential of biodiversity genomics, BMC genomics 24 (1):1-9.
Russell J Stewart, Paul B Frandsen, Steffen U Pauls, Jacqueline Heckenhauer (2022). Conservation of three-dimensional structure of Lepidoptera and Trichoptera L-Fibroins for 290 million years, Molecules 27 (18): 5945.
Jacqueline Heckenhauer, Paul B Frandsen, John S Sproul, Zheng Li, Juraj Paule, Amanda M Larracuente, Peter J Maughan, Michael S Barker, Julio V Schneider, Russell J Stewart, Steffen U Pauls (2022). Genome size evolution in the diverse insect order Trichoptera, GigaScience 11: giac011, https://doi.org/10.1093/gigascience/giac011.
Blanca Ríos-Touma, Ralph W Holzenthal, Ernesto Rázuri-Gonzales, Jacqueline Heckenhauer, Steffen U Pauls, Caroline G Storer, Paul B Frandsen (2022). De novo genome assembly and annotation of an Andean caddisfly, Atopsyche davidsoni Sykora, 1991, a model for genome research of high-elevation adaptations, Genome Biology and Evolution 14 (1): evab286, https://doi.org/10.1093/gbe/evab286.
Xuankun Li, Emily Ellis, David Plotkin, Yume Imada, Masaya Yago, Jacqueline Heckenhauer, Timothy P Cleland, Rebecca B Dikow, Torsten Dikow, Caroline G Storer, Akito Y Kawahara, Paul B Frandsen (2021): First annotated genome of a mandibulate moth, Neomicropteryx cornuta, generated using PacBio HiFi sequencing, Genome Biology and Evolution 13 (10): evab229, https://doi.org/10.1093/gbe/evab229.
Scott Hotaling, John S Sproul, Jacqueline Heckenhauer, Ashlyn Powell, Amanda M Larracuente, Steffen U Pauls, Joanna L Kelley, Paul B Frandsen (2021). Long reads are revolutionizing 20 years of insect genome sequencing, Genome Biology and Evolution 13 (8): evab138, https://doi.org/10.1093/gbe/evab138.
Lindsey K Olsen*, Jacqueline Heckenhauer*, John S Sproul, Rebecca B Dikow, Vanessa L Gonzalez, Matthew P Kweskin, Adam M Taylor, Seth B Wilson, Russell J Stewart, Xin Zhou, Ralph Holzenthal, Steffen U Pauls, Paul B Frandsen (2021). Draft genome assemblies and annotations of Agrypnia vestita Walker, and Hesperophylax magnus Banks reveal substantial repetitive element expansion in tube case-making Caddisflies (Insecta: Trichoptera). Genome Biology and Evolution, 13(3), evab013.
Jacqueline Heckenhauer, Steffen U. Pauls (2021). Faszinierende Unterwasserarchitekten. Natur Forschung Museum 151: 74-76.
Jacqueline Heckenhauer, Paul B Frandsen, Deepak K Gupta, Juraj Paule, Stefan Prost, Tilman Schell, Julio V Schneider, Russell J Stewart, Steffen U Pauls (2019). Annotated draft genomes of two caddisfly species Plectrocnemia conspersa CURTIS and Hydropsyche tenuis NAVAS (Insecta:Trichoptera). Genome Biology and Evolution 11 (12): 3445-3451. https://doi.org/10.1093/gbe/evz264