Our Research
Research in the Hiller lab revolves around a central question in genomics and evolutionary biology:
What is the genomic basis of phenotypic differences between species?
Our focus is explicitly on differences between species as opposed to differences within a species. We aim at discovering the genomic determinants of macroevolutionary trait differences, which is important to understand how nature’s spectacular phenotypic diversity has evolved. Our current focus is on studying the genomic basis of vertebrate traits that have potential applications in biomedicine or other fields.
To understand the molecular basis of biological diversity, we combine comparative genomics with genome sequencing and methods development. Our current research is interdisciplinary and includes
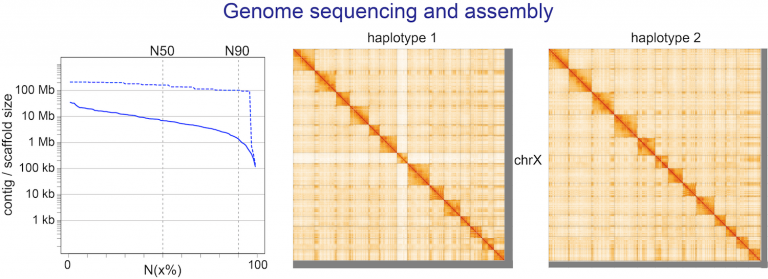
- sequencing reference-quality genomes and transcriptomes of selected species with interesting traits,
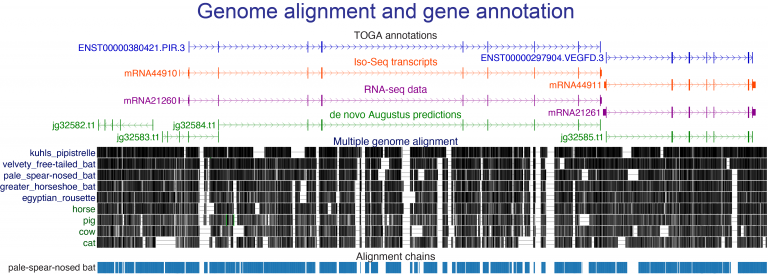
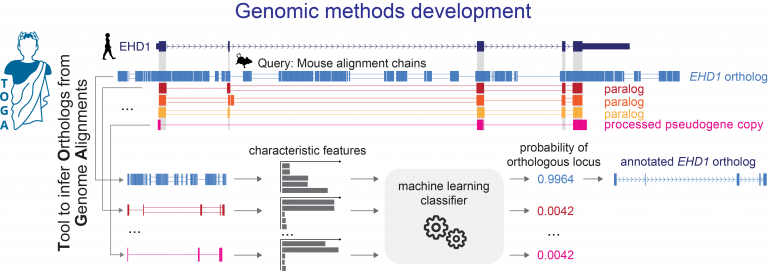
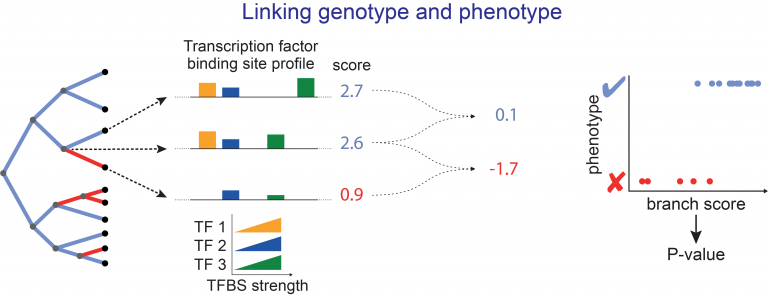
- developing new computational approaches to align genes and genomes, to annotate genes and infer orthologs, to accurately detect relevant evolutionary changes in functional genomic regions, and to discover associations between genotype and phenotype,
- running large-scale comparative genomic screens using existing and newly-sequenced genomes and our powerful methods toolbox to reveal the genomic basis of phenotypic adaptations and link phenotypic to genomic differences.
More information about our current projects:
- Genomic underpinnings of exceptional traits in bats
- Genomics of dietary adaptations
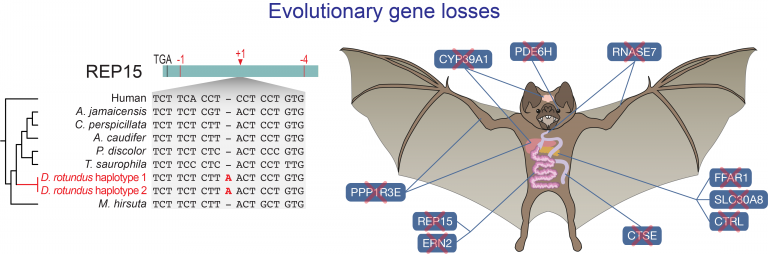
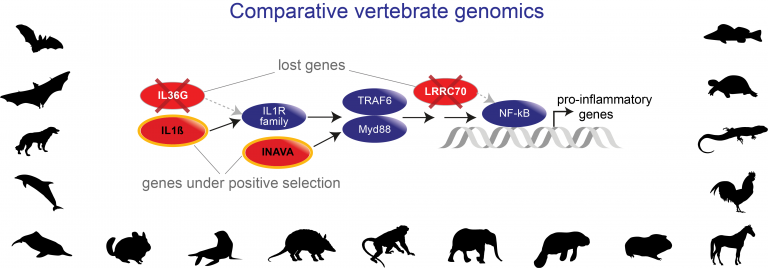
- Adaptive gene losses
- Forward Genomics: Linking phenotypic to genomic changes across species
- Development of comparative genomics methods
Videos
- Forward Genomics and linking phenotype to genotype (ESEB 2019; 10 min)
- Genomic determinants of adaptations in bats (PAG 2020; 26 min)
- Hat Evolution wirklich stattgefunden? (youtube in German)
Selected publications
- Morales AE, Dong Y, Brown T, Baid K, Kontopoulos DK, Gonzalez V, Huang
Z, Ahmed AW, Bhuinya A, Hilgers L, Winkler S, Hughes G, Li X, Lu P, Yang
Y, Kirilenko BM, Devanna P, Lama TM, Nissan Y, Pippel M, Dávalos LM,
Vernes SC, Puechmaille SJ, Rossiter SJ, Yossi Y, Prescott JB, Kurth A,
Ray DA, Lim BK, Myers E, Teeling EC, Banerjee A, Irving AT #, Hiller M #. Bat genomes illuminate adaptations to viral tolerance and disease resistance. Nature, https://doi.org/10.1038/s41586-024-08471-0, 2025 - Kirilenko BM, Munegowda C, Osipova E, Jebb D, Sharma V, Blumer M, Morales AE, Ahmed AW, Kontopoulos DG, Hilgers L, Lindblad-Toh K, Karlsson EK, Zoonomia Consortium, Hiller M #. Integrating gene annotation with orthology inference at scale. Science, 380 (6643), eabn3107, 2023
- Osipova E, Barsacchi R, Brown T, Sadanandan K, Gaede AH, Monte A, Jarrells J, Moebius C, Pippel M, Altshuler DL, Winkler S, Bickle M, Baldwin MW, Hiller M #. Loss of a gluconeogenic muscle enzyme contributed to adaptive metabolic traits in hummingbirds. Science, 379(6628), 185-190, 2023 pdf of article
- Blumer M, Brown T, Freitas MB, Destro AL, Oliveira JA, Morales A, Schell T, Greve C, Pippel M, Jebb D, Hecker N, Ahmed AW, Kirilenko BM, Foote M, Janke A, Lim BK, Hiller M #. Gene losses in the common vampire bat illuminate molecular adaptations to blood feeding. Science Advances, 8 (12), eabm6494, 2022
press and blogs are listed at altmetric - Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, Devanna P, Winkler S, Jermiin LS, Skirmuntt EC, Katzourakis A, Burkitt-Gray L, Ray DA, Sullivan KAM, Roscito JG, Kirilenko BM, Dávalos LM, Corthals AP, Power ML, Jones G, Ransome RD, Dechmann D, Locatelli AG, Puechmaille SJ, Fedrigo O, Jarvis ED, Hiller M #, Vernes SC #, Myers EW #, Teeling EC #. Six reference-quality genomes reveal evolution of bat adaptations. Nature, 583, 578–584, 2020
See also Nature Reviews Genetics highlight and Science feature. More press. - Huelsmann M, Hecker N, Springer MS, Gatesy J, Sharma V, Hiller M #. Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Science Advances, 5(9), eaaw6671, 2019
some press: New York Times, Science News, Discover Magazine, Smithsonian, Daily Herald, German Radio
F1000 recommended - Hecker N, Sharma V, Hiller M #. Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. PNAS, 116(8), 3036-3041, 2019
- Roscito JG, Sameith K, Parra G, Langer BE, Petzold A, Moebius C, Bickle M, Rodrigues MT, Hiller M #. Phenotype loss is associated with widespread divergence of the gene regulatory landscape in evolution. Nature Communications, 9:4737, 2018
- Lee JH, Lewis KM, Moural TW, Kirilenko B, Bogdanova B, Prange G, Koessl M, Huggenberger S, Kang C, and Hiller M #. Molecular parallelism in fast-twitch muscle proteins in echoloca