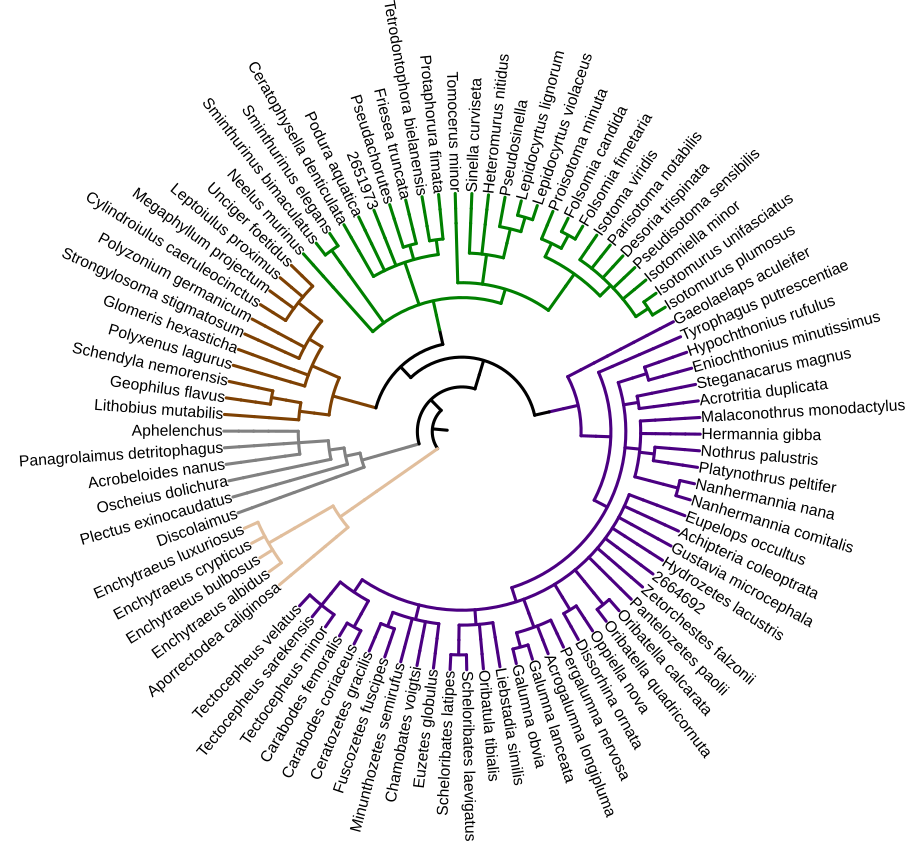Functional Environmental Genomics
Metagenomic monitoring of soil communities (MetaInvert)
Institution: Senckenberg Society for Nature Research
There is a tremendous diversity of invertebrates right under our feet: a single m2 soil in Germany might contain hundreds of species. Much of this diversity is unknown: it is estimated that for every known species there are about 20 that are still undescribed. These invertebrates are important contributors of ecosystem services essential for humanity, such as disease control, nutrient cycling, soil formation, or even primary production.
There are serious concerns that this diversity is rapidly lost. Since soil biodiversity loss likely impacts soil ecosystem services, there is demand from national and European authorities to understand and monitor soil biodiversity. However, extensive soil biodiversity monitoring is not yet feasible given the lack of methods that might allow to describe invertebrate community composition, biomass and functions diversity on a massive scale (Fig. 1). In MetaInvert we are developing the necessary genomic resources, laboratory and bioinformatic tools for a metagenomics-based monitoring of soil invertebrate communities. We also aim to start the standardisation process of the relevant approaches through ISO.

Fig. 1. Typical sample of soil microarthropods, containing hundreds of individuals from probably tens of species. Identification of such samples through microscopy requires highly trained specialists, and it is time consuming.
PROJECT tEAM
Project PIs
Miklos Bálint
Ricarda Lehmitz
Project members
Ina Schaefer
Andreas Kauk
Anna Küchler
Karin Hohberg
Alumni
Clément Schneider
Gemma Collins
Alexandra Schmidt
CONTRIBUTORS
Jörg Römbke (enchytraeids, ISO standardisation)
Rüdiger Schmelz (enchytraeids)
Pete Manning (BE-Spring)
Ulrich Burkhardt (Collembolas)
gENOME dATA
MetaInvert establishes a database of genomes for several hundred soil invertebrate species: springtails, oribatid mites, nematodes, potworms, myriapods, and several other groups. We already obtained genome sequences from over 120 species (Fig. 2). As of July 2020, over 100 additional species are processed for sequencing.
Fig. 2. Taxonomic relationships of over 80 soil invertebrates, genome-sequenced within the MetaInvert project. The tree reflects the project status as of May 2020. Species sequenced since then (~40) and species processed for sequencing are not included. The tree was generated with the Interactive Tree of Life tool
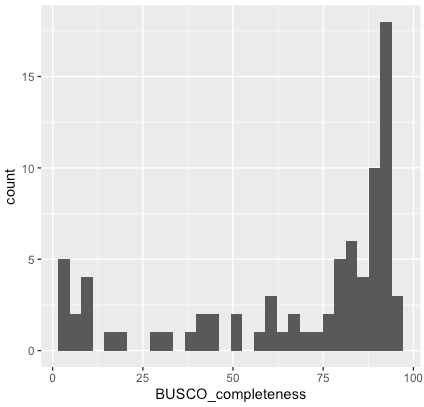
Genome data in MetaInvert is mostly generated from single individuals. Most of these individuals were collected and identified at the Soil Zoology Division, Senckenberg Museum of Natural History Görlitz. Most of the genomes are sequenced as Illumina short reads. The genomes have a coverage between 2x – 40x, and a varying completeness (Fig. 3). We are also experimenting with PacBio Sequel II for long-read sequencing. The MetaInvert database serves to improve species identification through shotgun metagenomics, to reveal evolutionary relationships, special adaptations, and host-microbiome associations.
Fig. 3. Completeness of the genomes of 80 soil invertebrate species sequenced until May 1, 2020, estimated with BUSCO counts
COOPEraTION PROJECTS
We performed a test project with mock communities to check the accuracy and efficiency of the shotgun metagenomic approach. This project constituted the recent MSc research of Alexandra Schmidt (Goethe University, Frankfurt) up until spring 2021. Mock communities were generated from a large variety of genome-sequenced species (oribatid and gamasine mites, nematodes, springtails, enchytraeids, tardigrades) at the Soil Zoology Division. Mocks were set up to allow for sensitivity and specificity analyses with regard to taxonomic identification and the recovery of biomass information from read numbers. The results show statistically significant association between biomass and shotgun sequencing read counts. However, taxonomic affiliation was the strongest predictor factor of reads. This hints at systemic taxonomic bias in DNA extraction efficiency, library preparation or sequencing.
The “BE-Spring: Discovering Collembola biodiversity on grasslands with emerging genomic and metagenomic tools” is a DFG-funded project within the Biodiversity Exploratories. It runs between 2020-2023, and constitutes the PhD research of Anna Küchler. BE-Spring complements the MetaInvert genome data with genomes of the most abundant soil springtails from the 150 Exploratories grassland plots to establish the Springtail Genome Reference Database (SGRD) of Germany. BE-Spring implements the shotgun metagenomic approach established within MetaInvert to understand
- how environmental drivers and the intensification of grassland use influence springtail community composition and function, and
- how springtails relate to other biological components of the grassland ecosystems
Group expertise / Methods
- environmental DNA
- Metabarcoding
- Soil arthropods collection
- Collembola taxonom
- Morphology
- Imagery: phase contrast and DIC highend microscope
- SEM
- Phylogenetics
- Python
- R
- machine learning
- metagenomics
- community ecology
SELECTED PUBLICATIONS
Schneider C, Woehle C, Greve C, et al (2021). Two high-quality de novo genomes from single ethanol- preserved specimens of tiny metazoans (Collembola). GigaScience.
Bálint M, Nowak C, Márton O, et al. (2018). Accuracy, limitations and cost efficiency of eDNA-based community survey in tropical frogs. Molecular Ecology Resources, 18(6), 1415–1426.